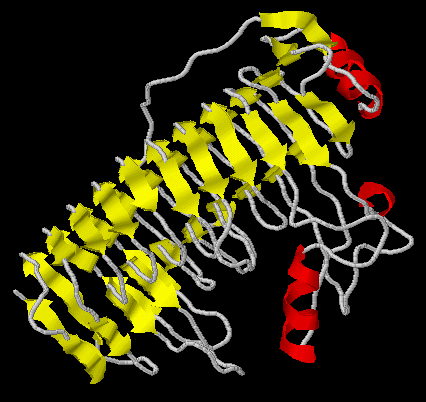
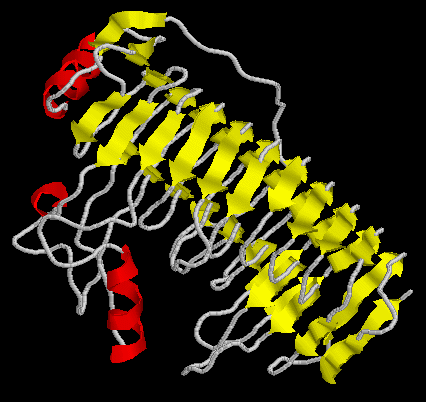
Alignment in FASTA format
Alignment No. 1: >3sil (the target sequence) ------------------------------------------------------------------------------ ---------------------------------------------------------------KSVVFKAEGEHFTD- ------QKGNTIVGSGSGGTT---------------KYFRIP------AMCTTSKGTIVVFADARHNTASDQSFIDT- --AAARSTDGGKTWNKKIAIYNDRVNSKLSRVMDPTCIVANI >1kit (the PDB template sequence) ALFDYNATGDTEFDSPAKQGWMQDNTNNGSGVLTNADGMPAWLVQGIGGRAQWTYSLSTNQHAQASSFGWRMTTEMKV LSGGMITNYYANGTQRVLPIISLDSSGNLVVEFEGQTGRTVLATGTAATEYHKFELVFLPGSNPSASFYFDGKLIRDN IQPTASKQNMIVWGNGSSNTDGVAAYRDIKFEIQGDVIFRGPDRIPSIVASSVTPGVVTAFAEKRVGGGDPGALSNTN DIITRTSRDGGITWDTELNLTEQINVSDEFDFSDPRPIYD-- Alignment No. 2: >jcha (the target sequence) MQQGQ-PMIILGEDSQLTSGQDAQSMNITAGKAVAEAVRTTLGPKGMDKMLVDSSGQVVV TNDGFTILKEMDIDHPAANMIVEVSETQETEVGDGTTTAVIIAGELLDQAEDLLESDVHA TTIAQGYRQAAEKAKEVLEDNAIEVTEDDHETLQKIAATAMTGKGAESAKDLLAELVVDA VLA---VKDDDSI-DTDNVSIEKVVGGTINNSELVEGVIVDKERVDENMPFAVEDADIAI LDDALEVRETEIDAEVNVTDPDQLQQFLDQEEKQLKEMVDQLVDVGADVVFVGDGIDDMA QHYLAKEGILAVRRAKSSDLKRLARATGGRVVSNLEDIEADDLGFAGSVGQKDIGGDERI FVEDVEDAKSVTLILRGGTEHVVDELERAIEDSLGVVRTTLEDGKVLPGGGAPETELSLT LREFADSVGGREQLAVEAFAEALDIIPRTLAENAGLDPIDSLVDLRSRHDGGEFAAGLDA YTGEVIDMEEEGVVEPLRVKTQAIESATEAAVMILRIDDVIAA >1a6dA (the PDB template sequence) MMTGQVPILVLKEGTQREQGKNAQRNNIEAAKAIADAVRTTLGPKGMDKMLVDSIGDIII SNDGATILKEMDVEHPTAKMIVEVSKAQDTAVGDGTTTAVVLSGELLKQAETLLDQGVHP TVISNGYRLAVNEARKIIDEIAEKSTDD--ATLRKIALTALSGKNTGLSNDFLADLVVKA VNAVAEVRDGKTIVDTANIKVDKKNGGSVNDTQFISGIVIDKEKVHSKMPDVVKNAKIAL IDSALEIKKTEIEAKVQISDPSKIQDFLNQETNTFKQMVEKIKKSGANVVLCQKGIDDVA QHYLAKEGIYAVRRVKKSDMEKLAKATGAKIVTDLDDLTPSVLGEAETVEERKIGDDRMT FVMGCKNPKAVSILIRGGTDHVVSEVERALNDAIRVVAITKEDGKFLWGGGAVEAELAMR LAKYANSVGGREQLAIEAFAKALEIIPRTLAENAGIDPINTLIKLKADDEKGRISVGVDL DNNGVGDMKAKGVVDPLRVKTHALESAVEVATMILRIDDVIAS |